ASTMH 2017, Sofonias Tessema: “Fine-scale population genetics of P. falciparum in Northern Namibia”
Countries: Namibia
Published: 08/11/2017
In collaboration with ASTMH, Image Audiovisuals, and session presenters, MESA brings you this webcast from the 66th ASTMH annual meeting in Baltimore, November 2017
Title: “Fine-scale population genetics of P. falciparum in Northern Namibia”
Speaker: Sofonias Tessema, University of California, San Francisco (UCSF)
Session information:
Symposium 0137: “Malaria: Genetics and Genomics”
Wednesday, 8 November, 10:15 – 12:00 PM, Convention Center – Room 321/322/323 (Level 300)
Abstract:
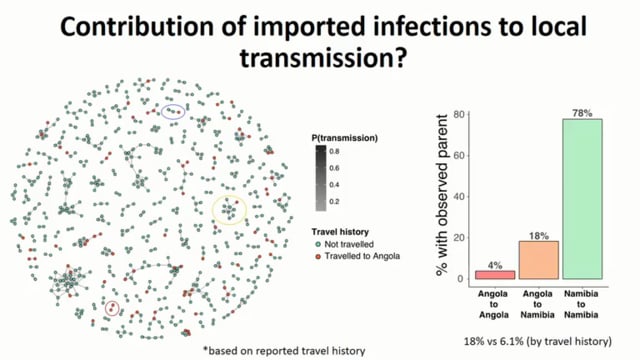
Namibia has a national goal to eliminate malaria by 2020, however, pockets of transmission and risks of importation still remain in Northern regions bordering Angola and Zambia. Molecular epidemiology can reveal important insights regarding contemporary and historical transmission and assess the impact of sustained control and elimination efforts. A total of 1696 samples from symptomatic malaria cases collected in 2016 from 23 clinics in 3 districts of the Kavango East region (Rundu (n=611), Nyangana (n=382), Andara (n=430)) and 6 clinics from the Zambezi region (n=273) were genotyped using 26 neutral microsatellite markers. 551 (32.5%) of the samples had monoclonal infections, of which 511 were unique haplotypes. Most infections in Kavango region contained multiple parasite genotypes (71%) with mean multiplicity of infection (MOI) of 2.4, high within-host diversity (mean Fws=0.7) and high genetic diversity (mean HE = 0.74), reflecting high rates of transmission and superinfection present during the regional outbreak which occurred at that time. Fine-scale temporal and spatial variation was observed in MOI consistent with the history of transmission intensity within this region. In contrast, Zambezi region had significantly lower polyclonal infection (49%), MOI (1.7), higher mean FWSscore (0.86) and lower genetic diversity (0.69), p < 0.001 vs. Kavango for all. In all districts, low but significant linkage disequilibrium (ISA: 0.01 – 0.04, p < 0.001) and genetic differentiation between districts (Gst: 0.02-0.06, p < 0.001) were observed. Genetic differentiation was strongly correlated with the distance between the districts (rho= 0.95, p = 0.003), reflecting some population fragmentation due to isolation by distance along with spread of parasites between the districts via vector or human travel. The findings of this study suggest targeted elimination of malaria in one region would be difficult to achieve without a similar effort throughout Northern Namibia. Integrated analyses of genetic and human mobility data can give insight to accurately estimate routes of parasite importation and transmission dynamics in space and time.
Fine-scale population genetics of P. falciparum in Northern Namibia
THEMES: Basic Science