ASTMH 2017, Oliver Watson: “Characterizing the potential bias within genomic tools for inferring changes in Plasmodium falciparum transmission intensities”
Collaborator(s): Imperial College London, United Kingdom
Published: 08/11/2017
In collaboration with ASTMH, Image Audiovisuals, and session presenters, MESA brings you this webcast from the 66th ASTMH annual meeting in Baltimore, November 2017
Title: “Characterizing the potential bias within genomic tools for inferring changes in Plasmodium falciparum transmission intensities”
Speaker: Oliver Watson, Imperial College London
Session information:
Symposium 0119: “Malaria: Advances in Modeling and Technology for Malaria”
Wednesday, 8 November, 8:00 – 9:45 AM, Convention Center – Ballroom I (Level 400)
Abstract:
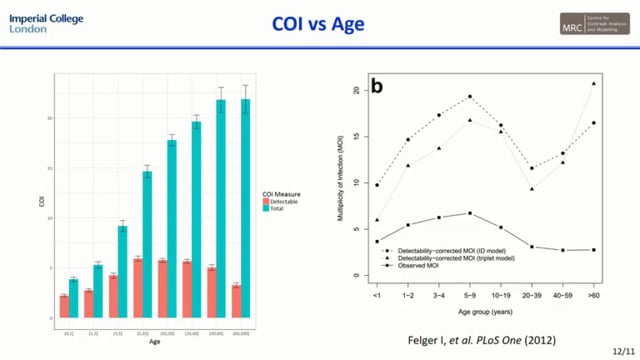
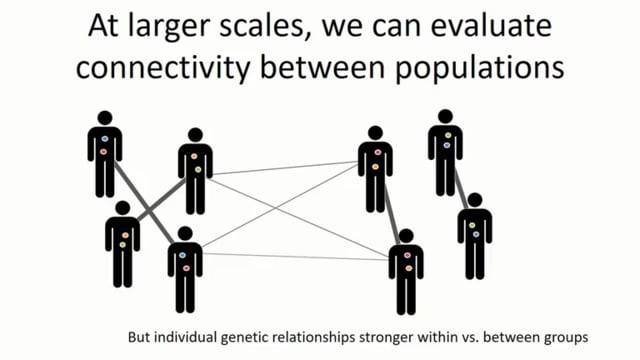
Substantial progress has been made globally to control malaria, however, there is a growing need for innovative new tools to ensure continued progress. In response, a number of genomic studies have been conducted detailing the potential for utilising measures of Plasmodium falciparum genetic diversity to infer changes in transmission intensity and the role of intervention policies. This work has revealed that the complexity of infection (COI), genetic relatedness and degree of clonality could be used as metrics with which to guide programmatic surveillance efforts. These studies, however, have noted a need to better understand both the interacting role of super-infection and cotransmission upon genetic epidemiological models and the impact of transmission heterogeneity within the population. Here we present a mathematical transmission model to identify potential bias that may be introduced by these processes upon the predictive accuracy of these genomic tools. Using an individual-based model of both human and mosquito dynamics, we incorporate a parasite genetic barcode model that identifies parasites based on 24 unlinked SNPs. Our modelling predicts firstly that heterogeneity within the human population can lead to bias in estimates of the mean population COI, with preferentially sampling clinical cases and young children leading to potential underestimates of transmission declines as a result of interventions. As a result, we identify optimal sampling schemes that ensure sufficient representation of the genetic diversity of the parasite population for accurate inferences of transmission intensity. Additionally, we characterise the impact of increased rates of cotransmission, and through sensitivity analysis identify transmission settings in which the role of cotransmission leads to overestimation of transmission intensity based upon genetic diversity inference. Through highlighting where potential bias exists, we further show support for how genomic tools could be deployed effectively to provide real-time inference upon the effectiveness of intervention programmes.
THEMES: Basic Science | Modeling