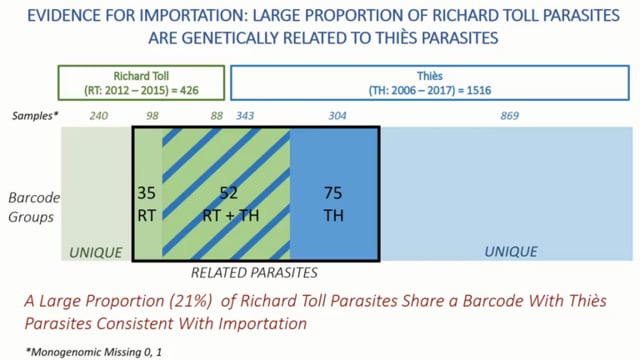
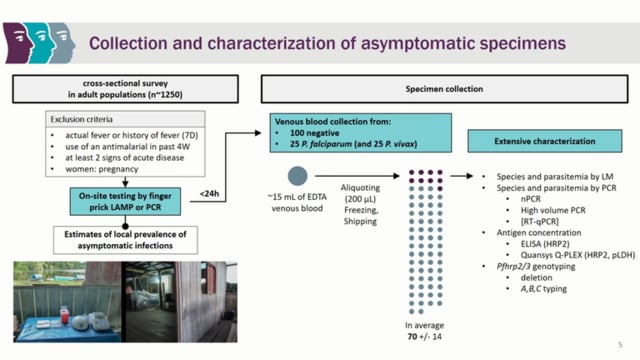
Molecular Approaches to Malaria (MAM) Conference – 2024: Day 1
Monday, 19th February 2024
Published: 19/02/2024
This report is brought to you by the MESA Correspondents Angela Rumaseb, Katelyn Stanhope, Mohini Anjna Shibu, and Nutpakal Ketprasit. Senior editorial support has been facilitated by Carla Proietti, Emma McHugh and Danushka Marapana.
THEMES: THEMES: Basic Science
MESA Correspondents bring you cutting-edge coverage from the MAM 2024 Conference “Malaria in a Changing World”.
Session 1 – Life Cycle Biology – pre-Erythrocytic
The conference kicked off with the keynote presentation by Photini Sinnis (John Hopkins University, USA). She spoke on the current paradigm that infected mosquitos are infectious by delving into the quantitative understanding of malaria transmission. Sinnis and her team used a mouse model to determine the likelihood of Plasmodium yoelli infection after exposure to infected mosquitoes. She presented the association between mosquito parasite burden, inoculum size and infection likelihood, and suggested that parasite numbers mattered for onward transmission. This association of mosquito sporozoite salivary gland load and infection rate is non-linear, with mosquitoes carrying over 10000 salivary gland sporozoites being more likely to transmit disease. She concluded by suggesting the possibility that as little as 20% of infected mosquitoes might account for a substantial 80% of the infections, suggesting the presence of super-spreader mosquitoes within the population.
Friedrich Frischknecht (Heidelberg University, Germany) delivered a comprehensive presentation on gametogenesis and parasite development in mosquitoes. He underscored the importance of some of the cytoskeleton proteins in gametes and elucidated how studying mosquito stages can offer potential intervention strategies. He highlighted the “upside-down” assembly of axonemes and the microtubule “push-pull mechanism”, crucial for gamete formation. He also emphasised the essential role of the microtubule interacting protein EB1 in nuclear segregation of gametes. Furthermore, Frischknecht and his team reported the essential role of a divergent Plasmodium actin-related protein 2/3 (Arp2/3) complex subunit, the ARPC1, in DNA segregation during male gametogenesis. They found that deletion of the actin binding protein profilin prevented mice infection. Frischknecht and his team were also able to generate 3D maps of oocyst maturation and sporozoite formation, providing valuable insights into the developmental processes of the parasite.
Hardik Patel (Seattle Children’s Hospital, USA) presented research on the liver stage of Plasmodium falciparum (P. falciparum). He discussed how blood stage host responses during concurrent infections can potentially suppress liver stage parasites, impacting the efficacy of live attenuated whole sporozoite vaccine. He reports that contrary to previous studies, the blood stage induced iron-regulating hormone hepcidin was not found to be responsible for liver stage development suppression. Instead, Patel observed that the host’s cytokine response, especially interferon gamma (IFNγ), played a pivotal role in suppressing the growth of the liver stage parasite, with this effect varying among different Plasmodium species. Furthermore, Patel reported that the effect of IFNγ resulted in an increase in GABARAPs (mediators of lysosomal fusion) as well as reactive oxygen species, but not nitric oxide production.
Liliana Mancio-Silva (Institute Paster, France) opened her presentation by addressing the current knowledge gaps and challenges associated with the dormant liver parasites, hypnozoites. Her talk focused on the application of single cell transcriptomics in Plasmodium vivax (P. vivax) hypnozoites. Mancio-Silva’s team used a bioengineered human microliver platform to culture patient-derived P. vivax parasites for transcriptional profiling. Dual scRNA-seq was used to analyse parasite and host transcripts in individual hepatocytes. Through their research, they identified potential biomarkers of hypnozoites based on ten genes. She also showed that hypnozoites keep the proteolytic activity to stay viable. Moreover, Mancio-Silva presented the findings of their transcription analysis, revealing that hypnozoite-infected hepatocytes are transcriptionally distinct, with two genesHLA-C and B2M, being notably expressed. The work gives insight into the mechanisms regulating P. vivax dormancy and may lay a foundation for the development of new vaccines and drugs.
Lauren Carruthers (University of Glasgow, Glasgow) explores and discusses the different gene expression across different life cycles, focusing on the ZINGER protein family. Carruthers shows that these proteins, characterised by 3x CCCH zinc finger domains, exhibit varying expression patterns throughout the parasite’s life cycle. Through knockout studies, Carruthers showed that ZINGER proteins play important roles in different developmental stages, including gametocyte, ookinete, and oocyst formation. This underscores the pivotal roles played by ZINGER proteins in regulating Plasmodium gene expression, thereby offering potential targets for controlling parasite development and malaria disease. Carruthers’ research advances understanding of gene regulation mechanisms with implications for future interventions aimed at tackling malaria.
Abhinay Ramaprasad (Francis Crick Institute, UK) focused on the malaria parasite’s asexual blood stages particularly emphasizing the essential genes during egress stage. While the DiCre system offers conditional gene manipulation, its scalability poses challenges. To overcome this hurdle, Ramaprasad introduced the SHIFTiKO method, which is a scalable knockout system for simultaneously querying the essential and biological function of multiple genes in the malaria parasite. Ramaprasad also discussed the use of pCas9-duo, which utilises two guides, thereby increasing the efficiency of the system. He highlighted the advantage of this method which facilitates high-throughput screening and promises accelerated discovery of critical genes across various stages of malaria development. This scalable strategy holds potential for transforming the pace of gene function discovery, advancing efforts towards combating malaria effectively.
Session 2 – Molecular Epidemiology & Population genetics
Isabella Oyier (Kenya Medical Research Institute – Wellcome Trust, Kenya) gave an insight into the parasite population and drug resistance in West Africa by integrating malaria molecular epidemiology into routine surveillance (IMMERSE). Oyier underscored the importance of surveillance of allele distribution and diversity, as this significantly impacts vaccine design and administration strategies. Oyier also discussed the parasite genetic diversity between asymptomatic and febrile infections, highlighting a higher degree of clonality in the asymptomatic infections. Oyier displayed the timeline of national antimalarial policy changes in Kenya and highlighted the drug resistance markers associated with the administered drugs. Longitudinal analysis in the Kilifi County Hospital revealed a decline in chloroquine resistance alleles post-chloroquine cessation, demonstrating the efficacy of temporal Malaria Molecular Surveillance (MMS). Oyier emphasised the advantage of amplicon sequencing from dried blood spot samples, now deployed nationwide in Kenya in collaboration with the National Malaria Control Programme (NMCP), enabling real-time monitoring of drug resistance.
Alexander W. Macharia (Kenya Medical Research Institute – Wellcome Trust, Kenya) presented the outcome of a recall-by-genotype study where β-thalassaemia carriers were matched with a control to determine the impact of β-thalassaemia on the invasion of red blood cells (RBCs) by Plasmodium falciparum (P. falciparum). When compared to controls, flow-cytometry assays revealed a reduction in invasion efficiency in β-thalassaemia heterozygotes. Macharia also explored whether this effect persisted in individuals co-inheriting α-and β-thalassemia traits and found that the effect of reduced infection was lost upon acquiring the α-thalassaemia trait. Results from RBCs surface proteins analysis showed differences between β-thalassaemia and normal RBCs, including increased basigin expression and reduced CD71 and CD49d. Macharia highlighted the importance of understanding these mechanisms to guide strategies for malaria elimination.
Amy Ibrahim (London School of Hygiene & Tropical Medicine – LSHTM, UK) delved into an overlooked malaria parasite, Plasmodium malariae (P. malariae) and the associated challenges, including low parasitemia infection rate and the absence of in vitro culture methods. Ibrahim introduced a selective Whole Genome Amplification (sWGA) method that was developed to address these obstacles. Additionally, she also highlighted the development of the first database for P. malariae genomics. Population genetics analysis revealed distinct genetic differences between Asian and African P. malariae isolates. Given the absence of in vitro culture methods for P. malariae, Ibrahim and her team developed a CRISPR-Cas9 model using P. knowlesi to assess pyrimethamine susceptibility, unveiling both sensitive and resistant P. malariae genotypes. The genomic analysis sheds light on P. malariae biology and drug resistance mechanisms.
Sarah Auburn (Menzies School of Health Research, Australia) highlighted challenges associated with eliminating Plasmodium vivax (P. vivax), including the parasite’s ability to form hypnozoites, the lack of effective treatment markers and the absence of reliable molecular markers for drug resistance. Auburn presented novel tools that measure the relatedness of parasites through identity-by-descent (IBD), aimed at deciphering the origin of recurrent infections as a proof-of-concept. She demonstrated the use of genomic data to deconvolve polyclonal infections using DEploid software, revealing improved data resolution and uncovering different relatedness characteristics. Another tool presented was a microhaplotype genotyping assay on the MiSeq platform that is suitable for use in malaria endemic-countries. Using this novel tool and time-to-event modelling, Auburn and her team aimed to classify recurrent infections in clinical trials. She concluded by highlighting the diverse range of questions that molecular surveillance tools can address to support the NMCP.
Yannick Höppner (Bernhard-Nocht Institute for Tropical Medicine, Germany) discussed how protection against malaria is dependent on acquired immunity, although sterile immunity is rarely achieved. The pathogenesis of malaria, caused by P. falciparum, is closely linked to the expression of var genes, which encode a protein called PfEMP1 on the parasite’s surface. This protein plays a crucial role in how the parasite interacts with human cells. Höppner demonstrated that var gene profiles differ between malaria naive individuals and those who have been previously exposed. Sample analysis from a longitudinal study identified that parasitemia cyclical peaks seen in P. falciparum infections were associated with sequential switching of expressed var genes. He also presented the correlation observed between antibodies against PfEMP1 and var gene expression, concluding that infections exhibit two distinct patterns: B-type infections persist until the end stage, while C-type infections are maintained but at the expense of parasite sequestration.
Nguyen Thanh Thuy Nhien (Oxford University Clinical Research Unit, Vietnam) opened by highlighting the burden of malaria cases in Vietnam as the country approaches elimination. Nhien showcased the evolution of molecular surveillance in Vietnam highlighting how genetic surveillance informs the decision-making of NMCP. Nhien informed the audience about how drug resistant phenotypes detected through genotyping during sentinel site monitoring are reported to the NMCP. Nhien also presented the increasing prevalence of artemisinin and piperaquine resistance throughout the years. She highlighted that the change in drug resistance trend coincided with drug treatment change and hence the importance of continuing molecular surveillance to track the drug’s effectiveness, the spread of drug resistance and the identification of parasite strains causing outbreaks.
Session 3 – Life Cycle Biology – Blood stage – 1
Yi–Wei Chang (University of Pennsylvania, USA) and his team used in situ cryo-electron tomography to study the rhoptry secretion system in related blood parasites such as Cryptosporidium parvum and Toxoplasma gondii. This system, which utilises a molecular ultrastructure called the rhoptry secretory apparatus (RSA), is essential for parasite invasion. Chang utilised cryo-electron tomography to examine the RSA in P. falciparum merozoites. Through subtomogram averaging of over a hundred images, his team revealed that the rhoptries have three distinct morphologies, determined by their interaction with the apical vesicle (AV). Additionally, he introduced cryo-confocal fLM and cryo-FIB-SEM which have been used to create stacks of images to study the surface of infected-red blood cells. He also suggested that rhoptries and AVs may develop from different biogenesis processes. He hypothesised that rhoptries evolved from a common ancestral structure similar to the trichocysts used by ciliates.
Kirk Deitsch (Cornell University, USA) elucidated how var genes express and switch in P. falciparum. His team used RNA-seq to study epigenetic switching. In their dataset, parasite populations switch between “high singles” (single var gene expression resulting in high binding parasites) and “low manys” (multiple var gene expression resulting in low binding parasites (a.k.a var-null parasites)). Results showed that var-null parasites were not recognised by antibodies and could cause asymptomatic infections. He also explained the availability of intracellular S-adenosylmethionine (SAM), a methyl group donor which could influence var genes to switch. In conclusion, he suggested a shifting bias in var genes wherein parasite populations express only a select 5-6 var genes every switch.
Emma Jones (University of Cambridge, UK) used optical tweezers to better understand the mechanism of P. falciparum merozoite attachment to the red blood cell surface. Using tweezers to detach merozoites from red cell membranes allowed the identification of the roles of different merozoite ligands in parasite-red cell attachment. Using a knockout model, the loss of MSP1 and GAP45 proteins did not reduce detachment forces, however, knockouts of EBA and RH proteins did reduce detachment force. The team explored how shaking speeds affected parasitic growth rates in culture, and determined a complex relationship between shaking speed and growth. To more accurately emulate invasion within blood vessels the team created a microfluidic device through which infected red blood cells travel and are visualised via fluorescence staining. By moving beyond the traditional static methods exploring invasion, this new tool expands our understanding of the invasion process in vivo.
Danny Wilson (The University of Adelaide, Australia) and team investigated the function of merozoite surface proteins (MSPs) across Plasmodium species due to their potential as vaccine candidates. Previous studies of MSP2 and MSP4 determined that these proteins are essential whereas MSP5 had been determined to be non-essential. To explore and challenge this, Wilson used CRISPR-Cas9 knockouts and found that loss of PfMSP2 simultaneously allowed parasite invasion as well as potentiated antibodies against Apical Membrane Antigen 1. Knock-down experiment results showed that MSP4 in P. falciparum was essential, but conversely, was not found to be important in merozoite invasion in P. knowlesi. Opposite results were found for MSP5, which was found to be essential for P. knowlesi but not for P. falciparum.
Ross Waller (University of Cambridge, UK) spoke of an evolutionary cell biology approach to parasitology. His group analysed the evolution of genes in common between species of the Myzozoa phylum such as Plasmodium and Toxoplasma, and addressed the question of “what has changed and what has stayed the same”. They used a spatial proteomic method called hyperplexed localization of organelle proteins by isotope tagging (hyperLOPIT) to study the cellular locations of new and old proteins. They identified Maurer’s clefts, the RBC membrane, and micronemes to be compartments that are evolving the fastest. Waller also suggested that the “spatial proteome” changes throughout the lifecycle of the parasite.
Vasant Muralidharan (University of Georgia, USA) presented studies on rhoptry neck protein 11 (RON11), which was shown to localise to the rhoptries. A knockdown of RON11, using the DOZI system, impacted parasite growth and merozoite invasion. However, the knockdown did not impact rhoptry secretions as the release of RAP1 into the infected cells was observable. Ultra-expansion microscopy revealed that RON11 knockdown merozoites only contained a single rhoptry. Muralidharan concluded that RON1 may not be essential for production of the first developing rhoptry but was required for biogenesis of its partner. To conclude he suggested that RON11 may also have an independent function in merozoite invasion that is less understood.
Tobias Spielmann (Bernhard Nocht Institute for Tropical Medicine, Germany) discussed PfEMP1 which is encoded by the multigene var family, which is canonically understood to have mutually exclusive expression. Spielmann introduced a novel Selection Linked Integration (SLI) system that allowed activation of a single PfEMP1 of interest and prevented switching. This was confirmed by the inclusion of an epitope tag (HA-tag) to detect the PfEMP1. Such modified parasite strains 3D7 and IT4 were used to study the binding to receptors such as CSA, CD36, ICAM-1, and EPCR. Biotinylation identification (BioID) with PfEMP1 as bait was used to identify new proteins that were important for cytoadhesion in infected parasites. The candidates identified are now being characterised using a modified SLI system.
This report is brought to you by the MESA Correspondents Angela Rumaseb, Katelyn Stanhope, Mohini Anjna Shibu and Nutpakal Ketprasit. Senior editorial support has been facilitated by Carla Proietti, Emma McHugh and Danushka Marapana.
Published: 19/02/2024
This report is brought to you by the MESA Correspondents Angela Rumaseb, Katelyn Stanhope, Mohini Anjna Shibu, and Nutpakal Ketprasit. Senior editorial support has been facilitated by Carla Proietti, Emma McHugh and Danushka Marapana.
THEMES: Basic Science