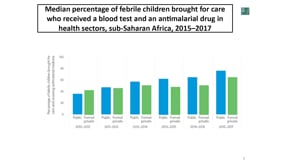
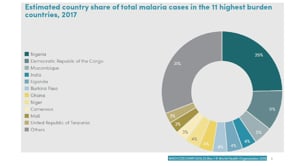
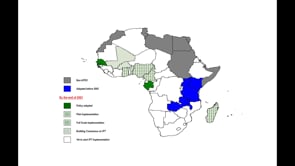
Last Updated: 02/12/2024
Evaluating the functional impact of genetic diversity on malaria vaccine candidates
Objectives
To use a reverse-vaccinology approach utilizing parasite genomic data directly from infected patients to identify and functionally interrogate the importance of antigen diversity in the development of highly effective malaria vaccines.
Malaria caused by Plasmodium falciparum remains one of the leading causes of death globally of both children and pregnant women. The recent global stall in the reduction of malaria deaths has made the development of a highly effective vaccine essential. A major challenge to developing an efficacious vaccine is the extensive diversity of Plasmodium falciparum antigens. While genetic diversity plays a major role in immune evasion and is a barrier to the development of both natural and vaccine-induced protective immunity, it has been underprioritized in the evaluation of malaria vaccine candidates. This proposal will use genomic approaches to credential next generation malaria vaccine candidates. Reverse vaccinology is a method of identifying potential antigens for a vaccine that starts with the genomic sequence of an organism and uses that information to identify epitopes and antigens that might make suitable vaccine candidates. Since the genome sequence of Plasmodium falciparum was published, only four new potential candidate vaccines have entered clinical development, including PfRh5. The main objective of the proposed study is to use a reverse-vaccinology approach utilizing parasite genomic data directly from infected patients to identify and functionally interrogate the importance of diversity in these antigens. For these current and novel candidates, including PfRh5 and binding partners, we will test the role of genetic diversity on immune neutralization by creating transgenic parasites by using efficient CRISPR-Cas9 genome editing. These parasite lines will be used to assess the role of specific variants in immune evasion prior to Phase 2 clinical trials. We will use IgG from malaria-immune individuals, followed closely in long-term longitudinal cohorts, and IgG from subjects in vaccine trials to assess the degree of inhibition of replication of malaria parasites by growth inhibition assays, neutrophil respiratory burst, and opsonophagocytosis of merozoites. This approach requires the cohesion of genomic sequencing technologies to identify potential candidate antigens and naturally occurring diversity, well-characterized human longitudinal cohorts to follow evolution of infection and immunity, standardized assays to serve as in vitro correlates of immunity, structure-based approaches for vaccine design, and strong ties to both scientists and institutions in endemic countries. The research team is uniquely positioned to combine these critical requirements to investigate the implications of parasite diversity on the development of protective immunity and vaccine efficacy, an essential factor to accelerate malaria vaccine discovery. This approach fills a critical need in the malaria vaccine development field in that it brings genetic diversity in candidate antigens to the forefront of vaccine candidate validation and credentialing. This study holds exceptional promise to discover new vaccine candidate combinations that will provide broadly neutralizing antibodies for inclusion in a globally effective vaccine, one that circumvents the parasite’s natural strategy to evade the immune system.
Sep 2022 — Jul 2027
$756,970