ASTMH 2017, Greg LaMonte: “Dual RNA sequencing identifies novel host biomarkers of Plasmodium hepatic infection”
Collaborator(s): University of California San Diego (UCSD), United States
Published: 08/11/2017
In collaboration with ASTMH, Image Audiovisuals, and session presenters, MESA brings you this webcast from the 66th ASTMH annual meeting in Baltimore, November 2017
Title: “Dual RNA sequencing identifies novel host biomarkers of Plasmodium hepatic infection”
Speaker: Greg LaMonte, University of California, San Diego (UCSD)
Session information:
Symposium 0137: “Malaria: Genetics and Genomics”
Wednesday, 8 November, 10:15 – 12:00 PM, Convention Center – Room 321/322/323 (Level 300)
Abstract:
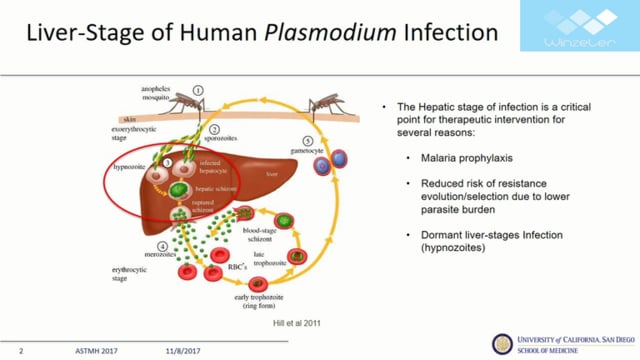
While efforts to ultimately eradicate malaria expand, the liver-stage of Plasmodium has gained in importance. As the first major step in the human cycle of infection, the liver-stage of Plasmodium infection represents the critical window for prophylactic intervention. The parasite burden in hepatic development is also substantially lower than in later human stages and therefore represents a critical developmental bottleneck in the parasite lifecycle. Several recent studies, using newly developed genetic tools, have identified host cell factors and receptors, such as EphA2, which play a critical role in parasite exoerythrocytic development.
In order to expand our understanding of liver-stage parasite biology, we report the findings of a dual RNA sequencing study to identify new host-parasite interactions via patterns of genetic dysregulation during hepatic infection. GFP-expressing Plasmodium berghei sporozoites were used to infect huh-7.5.1 liver cells, sorted via flow cytometry and combined host and parasite RNA was then extracted, sequenced and analyzed. We identified 3,142 host and 3,788 parasite genes with differential expression patterns during the Plasmodium exoerythrocytic lifecycle. These differentially expressed genes were validated through a variety of procedures, including comparison with a separate dual RNA-seq dataset from an independent human hepatocyte cell line (HC04), qPCR in multiple human cell lines as well as primary human hepatocytes and immunofluorescence in both P. berghei and P. vivax. Of all the differentially expressed genes identified, the most prominent was the host-factor mucin13 (muc13). We observe that muc13 is highly upregulated during late-stage parasite development in both P. berghei and P. vivax. In addition, genetic alteration of muc13, via either shRNA knockdown or CRISPR/Cas9 knockout, indicates that altered muc13 levels affect parasite development. We believe muc13represents a novel host biomarker of parasite infection, the characterization of which will increase our understanding of both parasite detection and hepatic development and advancing malaria eradication efforts.
THEMES: Basic Science