ASTMH 2017, Daniel Neafsey: “Comparative longitudinal population genomic surveys of Plasmodium falciparum malaria parasites in French Guiana and Thailand”
Collaborator(s): Harvard T.H. Chan School of Public Health (HSPH), United States
Countries: French Guiana, Thailand
Published: 08/11/2017
In collaboration with ASTMH, Image Audiovisuals, and session presenters, MESA brings you this webcast from the 66th ASTMH annual meeting in Baltimore, November 2017
Title: “Comparative longitudinal population genomic surveys of Plasmodium falciparum malaria parasites in French Guiana and Thailand”
Speaker: Daniel Neafsey, Harvard T.H. Chan School of Public Health
Session information:
Symposium 0137: “Malaria: Genetics and Genomics”
Wednesday, 8 November, 10:15 – 12:00 PM, Convention Center – Room 321/322/323 (Level 300)
Abstract:
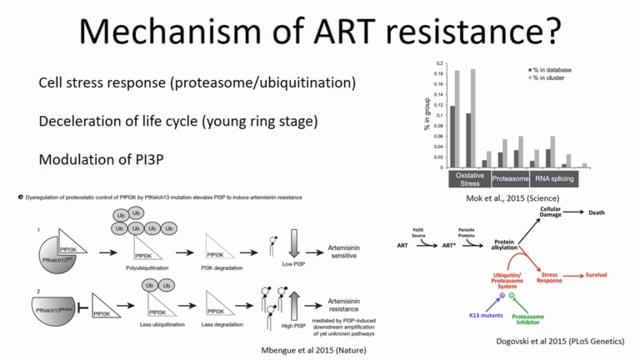
Drug-induced selection pressures combined with changing epidemiological patterns can create strong signals in population genomic datasets, especially when the selected variants are new mutations and when they only originate once. Conventional tests for selection on contemporaneous sample collections are less powered to detect selection on pre-existing variants, or selection that acts on particular combinations of variants in different parts of the genome. Longitudinal population genomic surveys can complement these deficiencies and reveal the pattern and process of resistance evolution. Artemisinin combination therapies (ACTs) are exerting selective pressure on P. falciparum in northern South America, a region where de novo resistance to previous antimalarial drugs has repeatedly emerged in the past. We sequenced the genomes of a longitudinal collection of 243 Plasmodium falciparum parasite samples from French Guiana spanning 17 years to identify mutations that have changed in frequency due to natural selection or genetic drift. We identified and annotated single nucleotide polymorphisms (SNPs) from Illumina sequencing data and evaluated the functional enrichment and classes of SNP alleles that increased or decreased in frequency over time. We also intersected these results with a previous study we performed of longitudinal population genomic variation in P. falciparum in Thailand, an epicenter of ACT resistance.In both locations we find evidence of changing allele frequencies in genes belonging to pathways previously implicated in artemisinin resistance (phosphatidylinositol phosphate regulation, proteasome/ubiquitination), as well as pathways that may represent an evolutionary response to drug pressure that does not explicitly confer drug resistance. We discuss the potential for this approach to detect early signs of ongoing adaptation and the implications of these findings for the mechanism of artemisinin resistance.
THEMES: Basic Science